This conference, presented by Genome Engineering and iPSC Center, has a lot of talented participants sharing their work through a virtual poster session. Check out the links below to see all of the unique research.
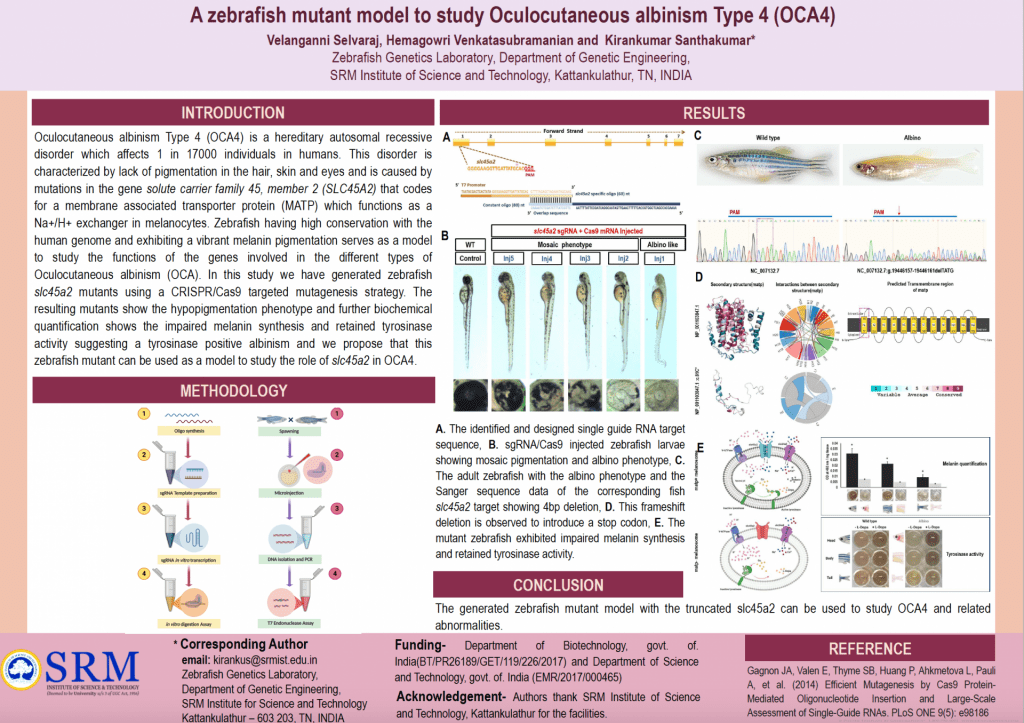
A zebrafish mutant model to study OCA4
Submitted by Velanganni Selvaraj , email: velu100170@gmail.com
Authors: Velanganni Selvaraj, Hemagowri Venkatasubramanian, Kirankumar Santhakumar
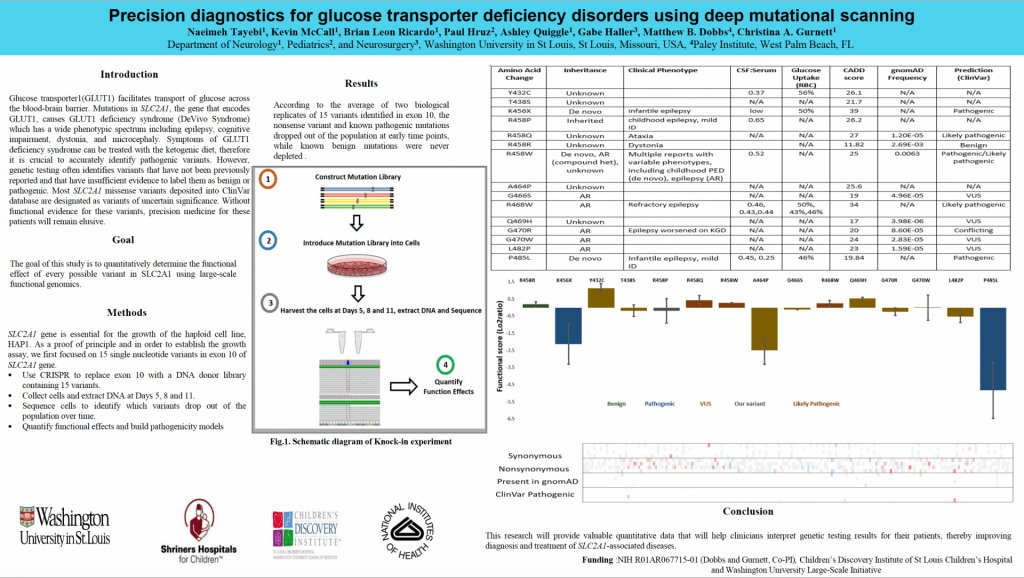
Paradigm shifting methods for high-throughput functional testing of all possible genetic variants: SLC2A1 and GLUT1 deficiency syndrome as a proof-of-principle
Submitted by Naeimeh Tayebi, email: ntayebi@wustl.edu
Authors:Naeimeh Tayebi, Kevin McCall, Brian Leon Ricardo, Paul Hruz, Ashley Quiggle, Gabe Haller, Matthew B. Dobbs, Christina A. Gurnett
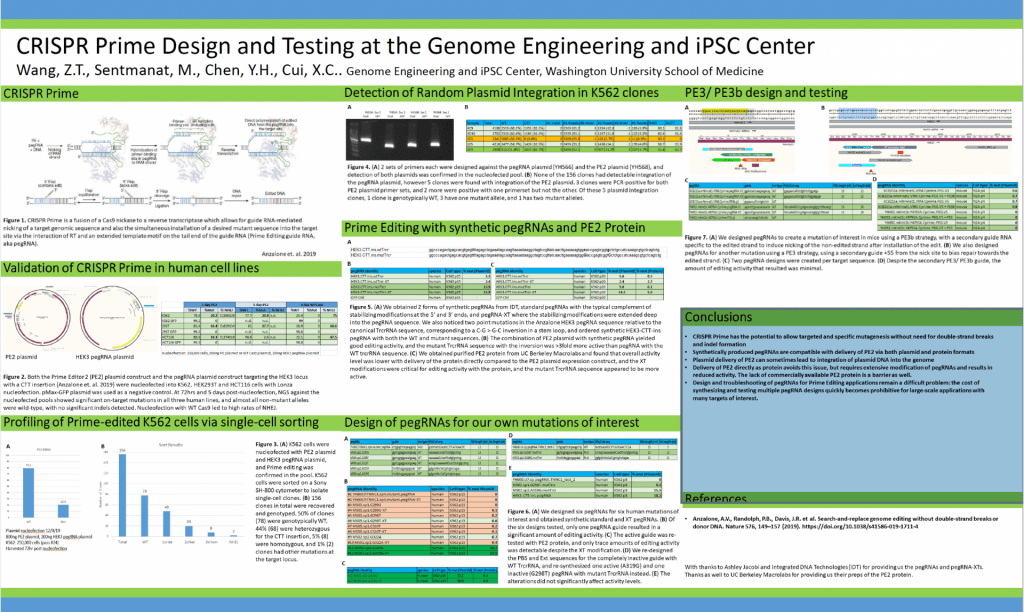
CRISPR Prime Design and Testing at the Genome Engineering and iPSC Center
Submitted by: Zi Wang, email: ztwang@wustl.edu
Authors: Wang, Z.T., Sentmanat, M., Chen, Y.H., Cui, X.C.
Hallmarks of Functional Genomics
Submitted by: William Buchser, email: wbuchser@wustl.edu
Author: William Buchser
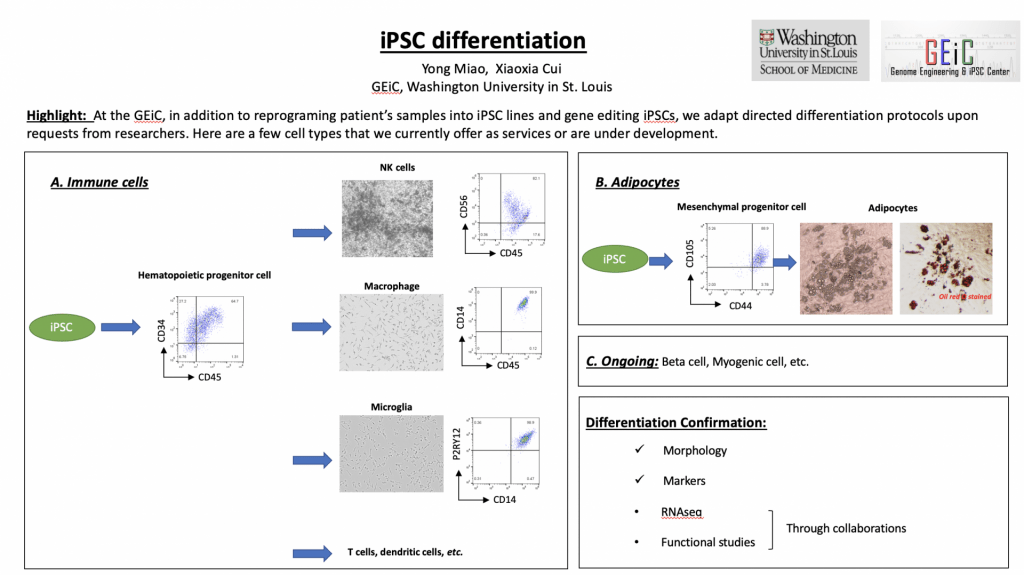
iPSC Differentiation
Submitted by Yong Miao, email: ymiao@wustl.edu
Authors: Yong Miao, Xiaoxia Cui
Did you miss the talks? Check out the recordings here.
Day 1
January 21, 2021 8 AM to 1 PM
- Charlie Gersbach, Ph.D., Rooney Family Associate Professor of Biomedical Engineering, Duke University
- Decoding and Programming the Genome with CRISPR Technologies
- Ibrahim Kurt, Ph. D. Candidate, Harvard University
- CRISPR C-to-G base editors for inducing targeted DNA transversions in human cells
- Traver Hart, Assistant Professor, Department of Bioinformatics and Computational Biology, The University of Texas MD Anderson Cancer Center
- CRISPR screening for functional genomics and cancer targeting
- Shengdar Tsai, Assistant Member of the Department of Hematology, St. Jude Children’s Research Hospital
- Understanding genetic and epigenetic effects on CRISPR-Cas9 genome-wide activity with CHANGE-seq
- Tim Peterson, Assistant Professor of Medicine, Washington University in St. Louis
- “Sphingolipids connect pathogens to aging-related disease”
- John Doench, Director R&D, Institute Scientist, Broad Institute of MIT and Harvard University
- “Massively parallel variant-to-function mapping with CRISPR base editor screens”
- Andrew Anzalone, Head of the Prime Editing Platform at Prime Medicine, Inc.
- “Prime Editing: Versatile and Precise Genome Editing without Double-strand Breaks or Donor DNA”
- Panel Discussion:
- Including Monica Sentmanat, Ph.D./Assistant Director of GEiC, Steven Hoffman Illumina, Cell & Molecular Biology Segment Manager, and other speakers from the event
- “Off targeting using nucleases, base editors, prime editors, and in screens”
Day 2
January 22, 2021 8 AM to 1 PM
- 8 AM Alejandro Chavez, Assistant Professor of Pathology & Cell Biology, Columbia University
- “Controlling gene expression with CRISPR-based tools”
- 8:30 AM Dongsheng Duan, Margaret Proctor Mulligan Professor in Medical Research, School of Medicine, University of Missouri, Columbia
- “CRISPR editing therapy for Duchenne muscular dystrophy”
- 9 AM Aron Geurts, Associate Professor, Department of Physiology, Medical College of Wisconsin
- Engineering Rats and Rat Model Resources
- 9:30 AM Xiaoxia Cui, Director of the Genome Engineering & iPSC Center, Washington University in St. Louis
- Reliable and Efficient Creation of Floxed Alleles in Mice
- 10 AM Andrew Yoo, Associate Professor of Developmental Biology, Washington University in St. Louis
- “Modeling late-onset neurodegenerative disorders via direct neuronal reprogramming of patient fibroblasts”
- 10:30 AM Martin Kampmann, Associate Professor, Institute for Neurodegenerative Diseases, UCSF Weill Institute for Neurosciences, University of California, San Francisco
- “CRISPR-based functional genomics in iPSC-derived models of brain disease”
- 11 AM. Luke Gilbert, Assistant Professor, Helen Diller Family Comprehensive Cancer Center, University of California, San Francisco
- “Genome-wide Programmable Transcriptional Memory by CRISPR-based Epigenome Editing”
- 11:30 AM Michael Kosicki, Postdoctoral Fellow, Lawrence Berkeley National Laboratory
- “Large deletions, small trouble”
- 12 PM Panel Discussion:
- Including Yi-Hsien Chen, Ph.D./Assistant Director of GEiC, Steven Hoffman Illumina, Cell & Molecular Biology Segment Manager, and other speakers from the event
- “On target large deletion, repression as KO alternative, off-targeting potential, continue first days”
