Defining Host-Donor Chimerism in Kidney Rejection by scRNA-seq

Dr. Andrew Malone
Dr. Malone’s team aim for this project was to define host and donor cell chimerism in kidney transplant biopsies by integrating scRNA-seq with de novo SNV calling, using parallel exome DNA sequencing to call host and donor variants. This aim was completed in full by using the services provided by MGI. Dr. Malone and the team used a total of 7 biopsy samples from 6 individuals and performed single-cell RNA sequencing using the 10X platform. Whole-exome sequencing was also performed on donor and recipient DNA from peripheral blood from each biopsy pair totaling 12 samples. The demuxlet computational tool was used to call the origin of each cell in each sample. There were no major deviations from the original methods for this aim. All analyses both biologic and bioinformatics were performed by the Malone and Humphreys team.
To complete the aim, the team, procured 7 kidney transplant biopsies from 6 patients undergoing cause biopsy. These included 2 non-rejecting biopsies and 5 biopsies with antibody-mediated rejection. They performed single-cell RNAseq using the 10X Genomics microfluidics platform about 20,000 cells per biopsy. These samples were sequenced to a depth of 50,000 reads. In parallel whole-exome sequencing was performed in donor and recipient DNA isolated from peripheral blood samples. A single .vcf file and BAM file from each biopsy were used as input for the demuxlet computational tool. The origin of each cell was called using this tool. Significant donor-recipient chimerism was found within the macrophage and lymphocyte populations. Analysis suggests that the donor-recipient ratio of macrophages is rejection status dependent whereas that of lymphocytes is dependent on time post-transplantation. They found that donor and recipient macrophages do not fit neatly into M1 or M2 phenotypes. Donor origin T cells differentially express oxidative phosphorylation genes when compared with recipient origin T cells suggesting they are in a quiescent state.
This project demonstrated the feasibility of harnessing genetic variation to determine whether immune cells in rejection are coming from the transplant recipient or from the donor. Dr. Malone and the team showed they can perform this type of analysis on a small complex tissue sample such as a kidney biopsy. These data highlighted the gene expression differences between immune cells of donor and recipient origin. They envisage that future studies will build on this work by studying a greater number of samples in various kidney transplant diseases. Ultimately the findings from this work increase our understanding of transplant rejection, a disease that lacks good diagnostic tools and treatments.
For continued educational resources by this team, read the publication:
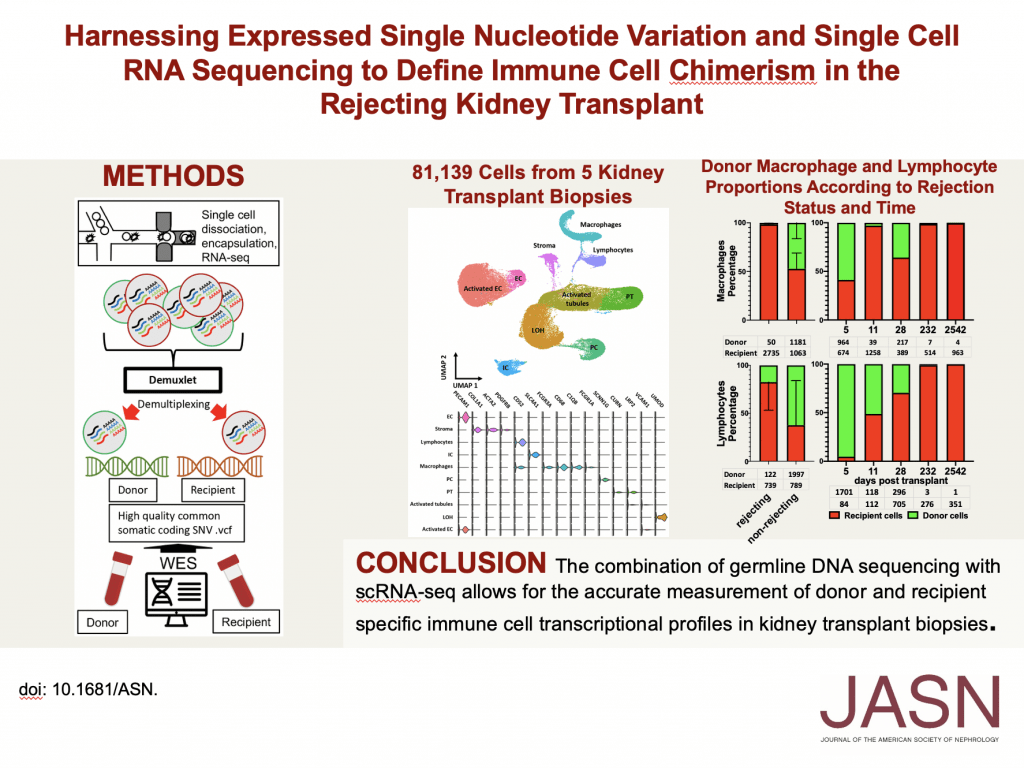
Harnessing Expressed Single Nucleotide Variation and Single Cell RNA Sequencing To Define Immune Cell Chimerism in the Rejecting Kidney Transplant, Andrew F Malone 1, Haojia Wu 1, Catrina Fronick 2, Robert Fulton 2, Joseph P Gaut 3, Benjamin D Humphreys 4 5, J Am Soc Nephrol, 2020 Sep;31(9):1977-1986. doi: 10.1681/ASN.2020030326. Epub 2020 Jul 15. Affiliations expand, PMID: 32669324, PMCID: PMC7461682 (available on 2021-09-01), DOI: 10.1681/ASN.2020030326
